Abstract
Introduction: Multiple gene expression profile assays have successfully prognosticated the overall survival (OS) outcomes of patients with Newly Diagnosed Multiple Myeloma (NDMM) and are now available for use in clinical practice. The genes included in these profiles encompass several aspects of cancer biology broadly such as cell-cycle biology, energy metabolism, migration, adhesion, immune evasion etc. Since altered energy metabolism is one of the hallmarks of all cancer cells, our goal was to take a deep-dive into specific genes associated with intracellular central carbon energy metabolism (CCEM) pathways such as oxidative phosphorylation (OxPhos) as well as glycolysis and lactate fermentation (Glyco-Lac) in MM cells to determine their impact on the pathogenesis and prognosis of MM patients.
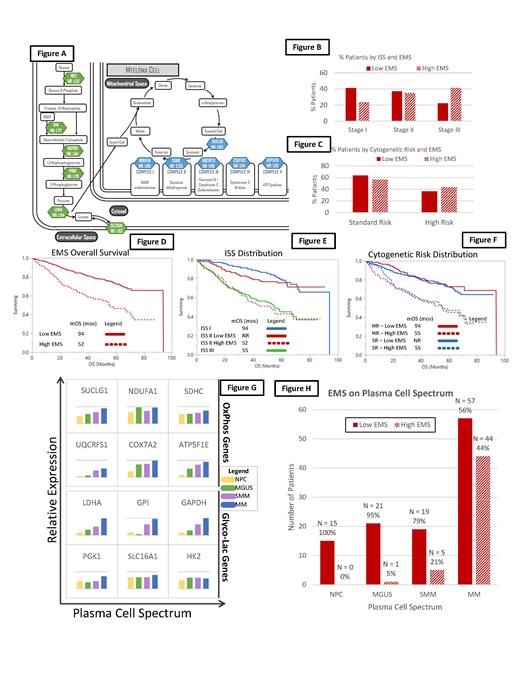
Methods: Gene expression level data via RNA sequencing from the COMMPASS dataset (made available by the Multiple Myeloma Research Foundation (MMRF)) was utilized. The expression levels of all genes associated with the OxPhos (N = 118) and Glyco-Lac (N = 34) pathways were evaluated in CD138+ cells derived from patients with NDMM. Each gene was dichotomized into low and high expression groups -based on their expression level above and below the median fragments per kilobase of transcript per million (FPKM), respectively. The prognostic significance of dichotomizing each of the 152 genes in this NDMM cohort was evaluated by levels of their hazard ratios (HRs) for OS determined by the log-rank method. Genes from the OxPhos and Glyco-Lac pathways were selected based on their significant negative impact on OS, wide representation of their biologic activity throughout the pathway and high independence from each other. This resulted in the selection of 12 distinct genes of interest. A scoring system consisting of assigning a point for every gene where their FPKM was above the median yielded a minimum of 0 and a maximum of 12 for the set of genes in the OxPhos and Glyco-Lac pathways to create an energy metabolism score (EMS). Patients were then categorized into "low" (0-8) or "high" (9-12) EMS groups. In addition, patients were designated as having high-risk cytogenetics if they had any of the following: t(4;14), t(14;16), t(14;20), del17p, and Amp 1q. Survival analysis was performed using the Kaplan-Meier survival analysis and compared via the log-rank method. Finally, the distribution of the EMS score among patients within a spectrum of plasma cell disorders such as, monoclonal gammopathy of undetermined significance (MGUS), smoldering multiple myeloma and MM as well as normal plasma cells was assessed using a previously published transcriptome dataset (GSE6477).
Results: A total of 766 patients were included in this study. The selected genes included in this scoring system and their biological relevance in the OxPhos and Glyco-Lac pathways are depicted in Figure A. The distribution of low and high EMS groups across ISS stage and cytogenetic risk are depicted in Figure B and C respectively. The median OS (mOS) and PFS for the high (N = 248) vs. low (N = 518) EMS groups was 55 vs. 94 months (HR: 2.41; (95%CI: 1.85-3.14); P <0.0001) (Figure D) and 24 vs. 42 months (HR: 1.74; (95%CI: 1.42-2.13); P <0.0001). Patients with ISS 2 stage disease categorized as having a high or low EMS score had a similar mOS to patients with ISS 3 and ISS 1 stage disease respectively (Figure E). Similarly, the mOS was similar among patients with standard risk and high-risk cytogenetics when categorized by low and high EMS (Figure F). Finally, the relative expression of most of the 12 select CCEM genes (Figure G) and the probability of a high EMS score increased among patients along the spectrum of plasma cell disorders (Normal plasma cell -> MGUS -> SMM -> MM) (Figure H).
Conclusions: The expression levels of specific genes associated with the activity of intracellular CCEM pathways such as glycolysis, lactate fermentation and oxidative phosphorylation in plasma cells provides evidence for the impact of cellular energy metabolism on the pathogenesis and survival outcomes of MM. This provides an opportunity for developing future studies aimed at understanding the energy metabolism pathways of plasma cell malignancies in greater detail so as to exploit them for diagnostic and therapeutic purposes in MM.
Kumar: Novartis: Research Funding; Amgen: Consultancy, Research Funding; Beigene: Consultancy; Carsgen: Research Funding; Merck: Research Funding; Antengene: Consultancy, Honoraria; Janssen: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Astra-Zeneca: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Oncopeptides: Consultancy; Roche-Genentech: Consultancy, Research Funding; KITE: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Takeda: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Tenebio: Research Funding; Bluebird Bio: Consultancy; Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; BMS: Consultancy, Research Funding; Abbvie: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Adaptive: Membership on an entity's Board of Directors or advisory committees, Research Funding; Sanofi: Research Funding. Dispenzieri: Alnylam: Research Funding; Sorrento Therapeutics: Consultancy; Oncopeptides: Consultancy; Pfizer: Research Funding; Takeda: Research Funding; Janssen: Consultancy, Research Funding. Kapoor: AbbVie: Research Funding; Takeda: Research Funding; Karyopharm: Consultancy; Cellectar: Consultancy; BeiGene: Consultancy; Pharmacyclics: Consultancy; Sanofi: Consultancy; Amgen: Research Funding; Ichnos Sciences: Research Funding; Regeneron Pharmaceuticals: Research Funding; Glaxo SmithKline: Research Funding; Karyopharm: Research Funding; Sanofi: Research Funding. Dingli: Apellis: Consultancy; Sanofi: Consultancy; Janssen: Consultancy; GSK: Consultancy; Novartis: Research Funding; Alexion: Consultancy. Lin: Gamida Cell: Consultancy; Takeda: Research Funding; Kite, a Gilead Company: Consultancy, Research Funding; Juno: Consultancy; Novartis: Consultancy; Legend: Consultancy; Janssen: Consultancy, Research Funding; Celgene: Consultancy, Research Funding; Bluebird Bio: Consultancy, Research Funding; Sorrento: Consultancy; Merck: Research Funding; Vineti: Consultancy. Gertz: Akcea Therapeutics, Ambry Genetics, Amgen Inc, Celgene Corporation, Janssen Biotech Inc, Karyopharm Therapeutics, Pfizer Inc (to Institution), Sanofi Genzyme: Honoraria; Aurora Biopharma: Other: Stock option; AbbVie Inc, Celgene Corporation: Other: Data Safetly & Monitoring; Ionis Pharmaceuticals: Other: Advisory Board; Akcea Therapeutics, Alnylam Pharmaceuticals Inc, Prothena: Consultancy. Fonseca: Aduro: Consultancy; Kite: Consultancy; Juno: Consultancy; Merck: Consultancy; Takeda: Consultancy; Caris Life Sciences: Membership on an entity's Board of Directors or advisory committees; Patent: Prognosticaton of myeloma via FISH: Patents & Royalties; AbbVie: Consultancy; Scientific Advisory Board: Adaptive Biotechnologies: Membership on an entity's Board of Directors or advisory committees; OncoTracker: Consultancy, Membership on an entity's Board of Directors or advisory committees; Mayo Clinic in Arizona: Current Employment; Amgen: Consultancy; BMS: Consultancy; Celgene: Consultancy; Bayer: Consultancy; Janssen: Consultancy; Novartis: Consultancy; Pharmacyclics: Consultancy; Sanofi: Consultancy; GSK: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal